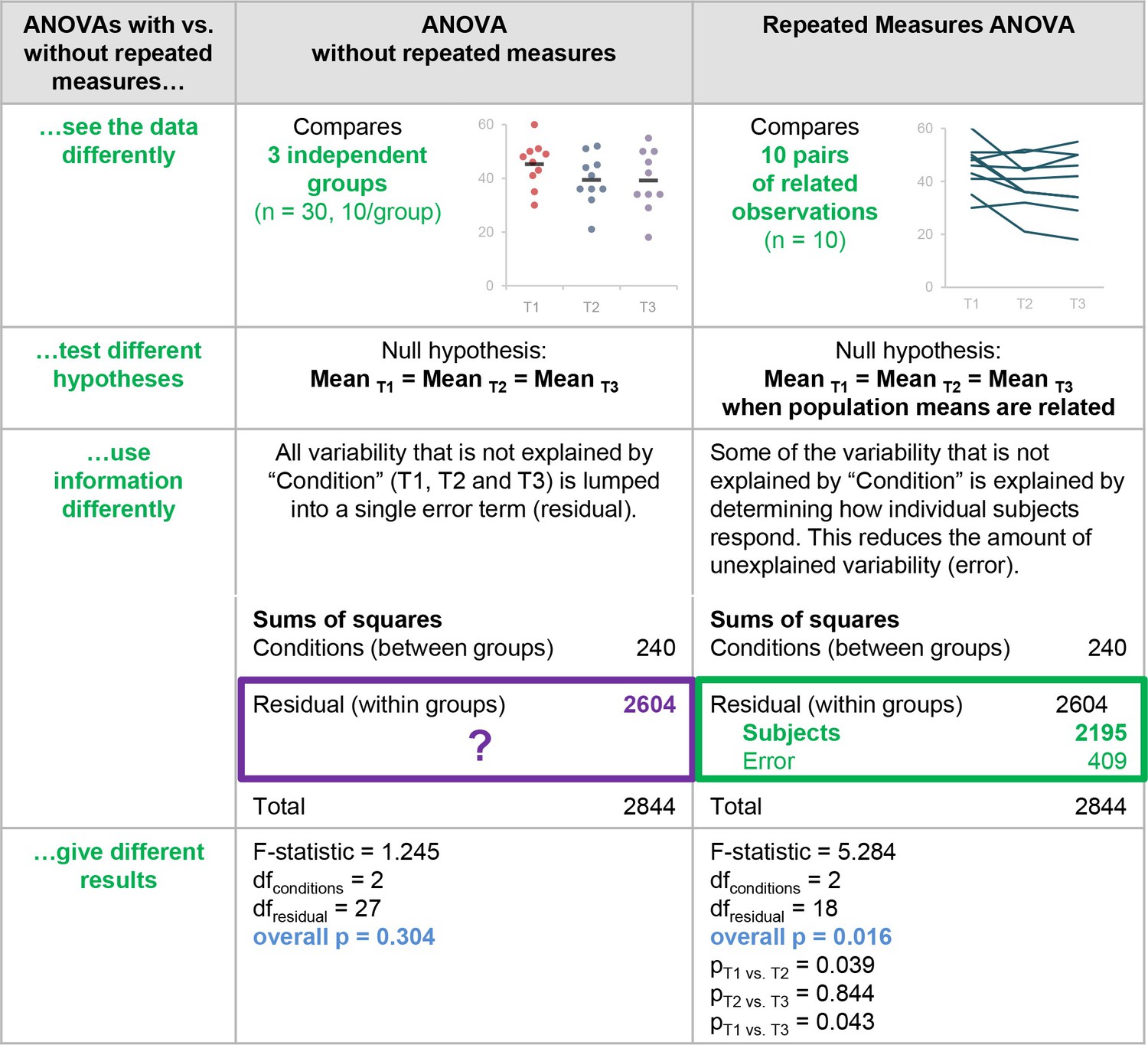
R Regressions
Linear regression
# Make some data # X increases (noisily) # Z increases slowly # Y is constructed so it is inversely related to xvar and positively related to xvar*zvar set.seed(0) x <- 1:20 + rnorm(20,sd=5) y <- 1:20 + rnorm(20,sd=5) type <- rep(LETTERS[1:2],each=10) # Make a data frame with the variables dat <- data.frame(x=x, y=y,type=type) # Show first few rows head(dat) |
## x y type ## 1 7.3147714 -0.1213394 A ## 2 0.3688332 3.8869782 A ## 3 9.6489963 3.6666818 A ## 4 10.3621466 8.0209475 A ## 5 7.0732072 4.7144661 A ## 6 -1.6997502 8.5180399 A |
scatterplot
plot(y ~ x, dat) # Add a regression line fitline <- lm(dat$y ~ dat$x) abline(fitline,col="red") lines(lowess(y, x), col = "blue") # lowess line (x,y) |
fit <- lm(y ~ x, dat) # fit <- lm(dat$y ~ dat$x) summary(fit) |
## ## Call: ## lm(formula = y ~ x, data = dat) ## ## Residuals: ## Min 1Q Median 3Q Max ## -8.967 -4.035 -0.081 5.481 8.297 ## ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 3.7511 2.6020 1.442 0.16659 ## x 0.6965 0.2143 3.250 0.00445 ** ## ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## Residual standard error: 3.046 on 30 degrees of freedom ## Multiple R-squared: 0.7528, Adjusted R-squared: 0.7446 ## F-statistic: 91.38 on 1 and 30 DF, p-value: 1.294e-10 |
l <- list(a = format(coef(model.lm)[1], digits = 4), b = format(abs(coef(model.lm)[2]), digits = 4), r2 = format(summary(model.lm)$r.squared, digits = 4), p = format(summary(model.lm)$coefficients[2,4], digits = 4)) eq <- substitute(italic(mpg) == a - b %*% italic(wt)~","~italic(R)^2~"="~r2~","~italic(P)~"="~p, l) ggplot(mtcars,aes(x=mpg,y=wt)) + geom_point(color = "blue") + stat_smooth(method = "lm",size = 2,color = "green",se=F) + geom_text(aes(x = 20, y = 1, label = as.character(as.expression(eq))), parse = TRUE) |
Logistic regression
A logistic regression is typically used when there is one dichotomous outcome variable (such as winning or losing), and a continuous predictor variable which is related to the probability or odds of the outcome variable. It can also be used with categorical predictors, and with multiple predictors.
Suppose we start with part of the built-in mtcars dataset. In the examples below, we’ll use vs as the outcome variable, mpg as a continuous predictor, and am as a categorical (dichotomous) predictor.
dat <- subset(mtcars, select=c(mpg, am, vs)) |
Continuous predictor, dichotomous outcome
If the data set has one dichotomous and one continuous variable, and the continuous variable is a predictor of the probability the dichotomous variable, then a logistic regression might be appropriate.
In this example, mpg is the continuous predictor variable, and vs is the dichotomous outcome variable.
# Do the logistic regression - both of these have the same effect. # ("logit" is the default model when family is binomial.) logr_vm <- glm(vs ~ mpg, data=dat, family=binomial) logr_vm <- glm(vs ~ mpg, data=dat, family=binomial(link="logit")) logr_vm |
## ## Call: glm(formula = vs ~ mpg, family = binomial(link = "logit"), data = dat) ## ## Coefficients: ## (Intercept) mpg ## -8.8331 0.4304 ## ## Degrees of Freedom: 31 Total (i.e. Null); 30 Residual ## Null Deviance: 43.86 ## Residual Deviance: 25.53 AIC: 29.53 |
summary(logr_vm) |
## ## Call: ## glm(formula = vs ~ mpg, family = binomial(link = "logit"), data = dat) ## ## Deviance Residuals: ## Min 1Q Median 3Q Max ## -2.2127 -0.5121 -0.2276 0.6402 1.6980 ## ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -8.8331 3.1623 -2.793 0.00522 ** ## mpg 0.4304 0.1584 2.717 0.00659 ** ## ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for binomial family taken to be 1) ## ## Null deviance: 43.860 on 31 degrees of freedom ## Residual deviance: 20.646 on 29 degrees of freedom ## AIC: 26.646 ## ## Number of Fisher Scoring iterations: 6 |
Multiple predictors with interactions
It is possible to test for interactions when there are multiple predictors. The interactions can be specified individually, as with a + b + c + a:b + b:c + a:b:c, or they can be expanded automatically, with a * b * c. It is possible to specify only a subset of the possible interactions, such as a + b + c + a:c.
This case proceeds as above, but with a slight change: instead of the formula being vs ~ mpg + am, it is vs ~ mpg * am, which is equivalent to vs ~ mpg + am + mpg:am.
# Do the logistic regression - both of these have the same effect. logr_vmai <- glm(vs ~ mpg * am, data=dat, family=binomial) logr_vmai <- glm(vs ~ mpg + am + mpg:am, data=dat, family=binomial) logr_vmai |
## ## Call: glm(formula = vs ~ mpg + am + mpg:am, family = binomial, data = dat) ## ## Coefficients: ## (Intercept) mpg am mpg:am ## -20.4784 1.1084 10.1055 -0.6637 ## ## Degrees of Freedom: 31 Total (i.e. Null); 28 Residual ## Null Deviance: 43.86 ## Residual Deviance: 19.12 AIC: 27.12 |
summary(logr_vmai) |
## ## Call: ## glm(formula = vs ~ mpg + am + mpg:am, family = binomial, data = dat) ## ## Deviance Residuals: ## Min 1Q Median 3Q Max ## -1.70566 -0.31124 -0.04817 0.28038 1.55603 ## ## Coefficients: ## Estimate Std. Error z value Pr(>|z|) ## (Intercept) -20.4784 10.5525 -1.941 0.0523 . ## mpg 1.1084 0.5770 1.921 0.0547 . ## am 10.1055 11.9104 0.848 0.3962 ## mpg:am -0.6637 0.6242 -1.063 0.2877 ## --- ## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ## ## (Dispersion parameter for binomial family taken to be 1) ## ## Null deviance: 43.860 on 31 degrees of freedom ## Residual deviance: 19.125 on 28 degrees of freedom ## AIC: 27.125 ## ## Number of Fisher Scoring iterations: 7 |
Compare
set.seed(1001) y = rnorm(1000) x = matrix(rnorm(10000000), 1000, 10000) system.time(for (i in 1:ncol(x)) lm.fit(y = y, x = cbind(rep(1,NROW(x)),x[,i]))$coefficients[2]) # user system elapsed # 3.661 0.000 3.662 system.time(cor(y,x)*sd(y)/apply(x,2L,sd)) # user system elapsed # 1.173 0.011 1.184 system.time({mx=colSums(x); (y%*%x-mx*mean(y))/(colSums(x*x)-mx*mx/length(y))}) # user system elapsed # 0.173 0.012 0.186 |
ad
B = 100000 g = 9.8 ## meters per second n = 25 tt = seq(0,3.4,len=n) ##time in secs, t is a base function X = cbind(1,tt,tt^2) A = solve(crossprod(X))%*%t(X) set.seed(1) betahat = replicate(B,{ y = 56.67 - 0.5*g*tt^2 + rnorm(n,sd=1) betahats = -2*A%*%y return(betahats[3]) }) sqrt(mean( (betahat-mean(betahat) )^2)) #sqrt(var(betahat)) |
## [1] 0.4297449 |