✨ Update Infos
2017-10-15: I thought my python3.6 on my Win7 was corrupted, so I reinstalled all my python packages.
But why not use Anaconda for windows instead, so let us first download it.
2018-10-7 : update using Win10 and python3.7.
2020-11-21: update python to 3.7.8
Note : The commands as follows was implemented by CMD / PowerShell (Make sure anaconda has been in your system environment) OR using anaconda-navigator ( Version 1.8.3 ), graphical interface to use conda.
1 2 3 4 5 6 7 8 9 10 11 12 conda update anaconda # update anaconda conda update conda # update conda or conda update -n base -c defaults conda conda update python # update python # Solving environment: done # All requested packages already installed. conda -V # 4.5.11 python -V # Python 3.7.0 python -m pip install --upgrade pip pip -V # pip 18.1 from d:\anaconda3\lib\site-packages\pip (python 3.7)
or you can install package locally:
conda install --offline --verbose C:\Users\Administrator\Desktop\qt-5.9.7-vc14h73c81de_0.tar.bz2
Packages Mirror in china
From USTC: http://mirrors.ustc.edu.cn/
Form TUNA: https://mirror.tuna.tsinghua.edu.cn/
1 2 3 4 5 6 7 8 9 10 11 12 # # conda config --add channels ... # conda config --remove channels ... conda config --add channels "http://mirrors.ustc.edu.cn/anaconda/cloud/bioconda/" conda config --add channels "http://mirrors.ustc.edu.cn/anaconda/cloud/conda-forge/" conda config --add channels "http://mirrors.ustc.edu.cn/anaconda/cloud/msys2/" conda config --add channels "http://mirrors.ustc.edu.cn/anaconda/pkgs/free/" conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/pkgs/free/ conda config --add channels https://mirrors.tuna.tsinghua.edu.cn/anaconda/pkgs/main/ conda config --add channels "defaults" # highest priority conda config --get channels # get the currnet channels .condarc conda config --set show_channel_urls yes
Conda itself can :
Query and search the Anaconda package index and current Anaconda installation.
Create new conda environments.
Install and update packages into existing conda environments.
Tips: --foobar and -f are equal args
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 # conda info -h conda search -h conda install -h conda update -h conda list -h # conda create -n puppies python conda activate puppies conda env list # # # base * D:\Anaconda3 # puppies D:\Anaconda3\envs\puppies conda install scipy scrapy # ... # The following NEW packages will be INSTALLED: # cssselect: 1.0.3-py37_0 defaults # parsel: 1.4.0-py37_0 defaults # pydispatcher: 2.0.5-py37_1 defaults # pytest-runner: 4.2-py37_0 defaults # queuelib: 1.5.0-py37_0 defaults # scrapy: 1.5.1-py37_0 defaults # w3lib: 1.19.0-py37_0 defaults # ... # Preparing transaction: done # Verifying transaction: done # Executing transaction: done conda list -n puppies
1 2 3 4 5 6 7 8 9 10 11 # packages in environment at D:\Anaconda3\envs\puppies: # # Name Version Build Channel certifi 2018.10.15 py37_0 defaults pip 10.0.1 py37_0 defaults python 3.7.0 hea74fb7_0 defaults setuptools 40.4.3 py37_0 defaults vc 14.1 h0510ff6_4 defaults vs2015_runtime 14.15.26706 h3a45250_0 defaults wheel 0.32.2 py37_0 defaults wincertstore 0.2 py37_0 defaults
1 2 3 4 conda create -n bunnies --clone puppies conda install -n bunnies jupyter scipy pandas matplotlib pillow redis psycopg2 pymysql beautifulsoup4 pyquery selenium scrapy flask django autopep8 pip install you-get conda list -n bunnies
1 2 3 4 # packages in environment at D:\Anaconda3\envs\bunnies: # # Name Version Build Channel ...
1 2 3 4 5 6 7 8 9 10 11 12 13 conda create -n tensorflow pip python=3.6 #tensorflow do not support py3.7 coz of async conda install -n tensorflow tensorflow conda info -e # conda environments: # # base * D:\Anaconda3 # bunnies D:\Anaconda3\envs\bunnies # puppies D:\Anaconda3\envs\puppies # tensorflow D:\Anaconda3\envs\tensorflow conda remove -n puppies --all conda activate bunnies pip freeze > lists.txt # export current python environment pip install -r lists.txt # import current python environment
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 # lists.txt jupyter ipykernel>=4.8.2 # for crawler requests aiohttp httpie beautifulsoup4 lxml html5lib>=1.0.1 pyquery selenium tesseract # tesserocr pillow tornado pyspider scrapy scrapy-redis ### twisted use conda instead # for databases redis psycopg2 pymysql # for ml numpy pandas matplotlib scipy pillow scikit-learn nltk jieba keras bleach>=2.1.3 tensorflow>=1.7.0 # for life py2exe you-get pyexcel # for web flask django # for style pycodestyle PyYAML # for Cipher pycryptodomex # windows
Learn conda in 30min : https://bit.ly/tryconda
Conda cheat sheet : https://conda.io/docs/_downloads/conda-cheatsheet.pdf
Bioconda resources :https://bioconda.github.io/recipes.html#recipes
Warning : This chapter was the archive from 2017-10-15 on Ubuntu 17.10, so the content is likely to be out of date.
Bioconda is a channel for the conda package manager specializing in bioinformatics software.
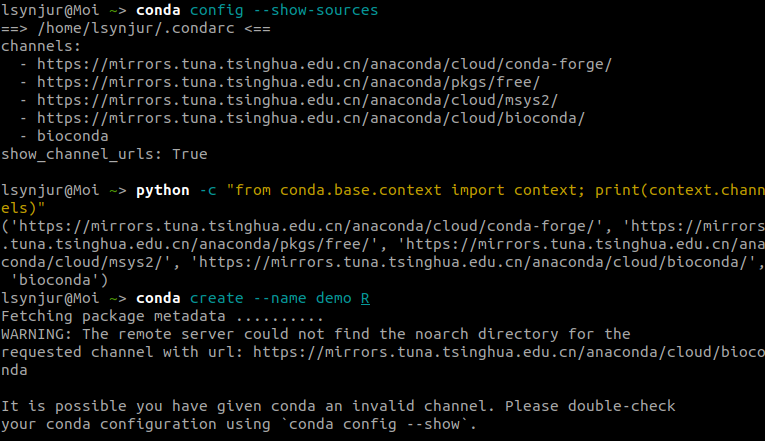
1 2 3 4 5 6 7 8 9 gedit .condarc channels: - https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/conda-forge/ - https://mirrors.tuna.tsinghua.edu.cn/anaconda/pkgs/free/ - https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/msys2/ - https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/bioconda/ - r - bioconda show_channel_urls: true
But when I trying to use TUNA bioconda mirror, I got an error!!!
https://mirrors.tuna.tsinghua.edu.cn/anaconda/cloud/bioconda/ mirror did not work properly,
but it exactly has noarch dir and soso, and you know the internet situations here in china, thus I want to build a local bioconda mirror.
Note : This chapter was the archive from 2017-10-15 on Ubuntu 17.10.
Download all files from bioconda mirror except the prefixes including bioconductor or r or ucsc first
1 2 3 4 5 conda index linux-64 cp -fu linux-64/repodata.json ./noarch bzip2 -k noarch/repodata.json
1 conda search -c file:///media/lsynjur/Doppelganger/bioconda --override-channels
–override-channels search the .condarc channels and the defaults channels , is that a bug?
1 2 3 conda env create -n bioc -f demo.yml conda env export -n bioc -f bioc.yml source activate bioc